This module defines the fish and its components. The model is discrete, time is based on time steps, in s. More...
Data Types | |
| type | history_array_fish |
| This type defines history arrays that are saved for the FISH class. More... | |
| type | fish_skel |
| This defines the root type for the FISH class. More... | |
| interface | smr |
| Calculate SMR, with generic interface to non-object oriented procedure. More... | |
| interface | stress_factor |
| Calculate stress pattern based on cubic spline interpolation over the grid defined by commondata::global_stress_factor_hour and commondata::global_stress_fact_suppress. More... | |
| type | food_item_eaten |
| type | food_item_eaten_mg |
| type | stomach |
| Defines the stomach of the fish. More... | |
| type | midgut |
| Defines the midgut of the fish. More... | |
| type | fish |
| Defines an umbrella class for the complete fish organism An outline of the ::fish class inheritance is presented below. More... | |
Functions/Subroutines | |
| elemental real(srp) function | stomach_vol (mass) |
| Calculate the stomach volume (ml) based on the fish body mass (g) using Salgado, A., Salgado, I., & Valdebenito, I. (2018). A direct and straightforward method for measurement real maximum fish stomach volume to improve aquaculture feeding research. Latin American Journal of Aquatic Research, 46(5), 880–889. https://doi.org/10.3856/vol46-issue5-fulltext-3. More... | |
| elemental subroutine | fish_init_new_zero (this, id_num) |
| Initialise an empty ::fish object to an empty zero state. More... | |
| pure real(srp) function | fish_smr_temp_bodymass_evan1990_scalar (this, temperature, is_per_hour, time_step) |
| SMR calculation from Evans 1992 data, scalar version. More... | |
| pure real(srp) function | fish_smr_temp_bodymass_evan1990_value (body_mass, temperature, is_per_hour, time_step) |
| SMR calculation from Evans 1992 data, non-object scalar version. More... | |
| elemental real(srp) function | amr_swim_basic (fish_mass, swimming_speed, time_unit) |
| Calculate the active metabolic rate of fish in units of oxygen consumption per hour. More... | |
| elemental real(srp) function | fish_amr_locomotion_energy_cost (this, temperature, is_exclude_smr, is_per_hour) |
| Calculate the active metabolic rate. This can be based on amr_swim_basic() or def_salmon::salmon_oxygen_consumption(). More... | |
| elemental real(srp) function | fish_whole_body_energy_trout (this) |
| Calculate the whole body energy content. More... | |
| elemental real(srp) function | energy2mass (body_energy) |
| Calculate the body mass from whole body energy. More... | |
| elemental subroutine | food_item_eaten_set_zero_init_state (this, mass, start_mass, delay) |
| Initialise an empty food item as ingested by the fish. Cleanup and set zero state. More... | |
| elemental subroutine | food_item_eaten_mg_set_zero_init_state (this, mass, start_mass, delay) |
| Initialise an empty food item as ingested by the fish. Cleanup and set zero state. More... | |
| elemental subroutine | food_item_eaten_eat_init_at_ingest (this, food_item_in, ingest_delay) |
| Initialise a single food item as it is ingested by the fish. Note that the ingested food item is copied from the external food item environ::food_item that comes as the input argument. More... | |
| elemental subroutine | food_item_eaten_update_age_in_stomach (this, t_increment) |
| This procedure updates the time the food item spent in the stomach of the fish. More... | |
| elemental subroutine | food_item_eaten_check_destruct (this, is_destructed, tolerance, max_time) |
| Check if the food item mass shrunk to near-zero and destruct it. More... | |
| elemental subroutine | food_item_eaten_mg_check_destruct (this, is_destructed, tolerance, max_time) |
| Check if the food item has been processed for more than commondata::global_maximum_duration_midgut s time in the midgut. More... | |
| elemental logical function | food_item_eaten_mg_exceed_max_is_to_destruct (this, max_time) |
| Check if the food item in the midgut has spent for more than the maximum time in the midgut after full absorption and should be destructed and evacuated (TRUE). More... | |
| elemental logical function | food_item_eaten_is_absorbed_full (this, max_absorb) |
| Check if the food item has been absorbed to the maximum possible mass. More... | |
| elemental real(srp) function | st_food_item_mass (time_in_process, dry_mass, ingestion_delay, water_uptake, rescale) |
| This function defines the mass of a single food item as it passes through the fish stomach. More... | |
| elemental subroutine | fish_stomach_init (this, mass) |
| Initialise an empty fish stomach object. More... | |
| impure subroutine | fish_stomach_update_step (this) |
| Update one time step of the fish stomach life cycle. More... | |
| elemental subroutine | fish_ingest_food_item (this, food_item_ingested) |
| This procedure shows what happens when the fish eats a single food item. More... | |
| elemental real(srp) function | fish_stomach_total_mass_food (this, tolerance) |
| Calculate the total mass of food in the fish stomach. More... | |
| elemental real(srp) function | fish_appetite_factor_stomach (this, fcapacity) |
| Calculate the fish appetite factor based on the relative stomach fullness. Appetite factor is equivalent to the probability of consuming any food item. More... | |
| elemental subroutine | fish_midgut_init (this, mass) |
| Initialise an empty fish midgut object. More... | |
| impure subroutine | fish_midgut_update_from_stomach (this, dry_mass, t_increment) |
| This procedure processes the food item in the midgut of the fish. More... | |
| elemental subroutine | fish_energy_balance_update (this, temperature, start_value) |
| Calculate and update the current energy balance of the fish. Note that the energy balance is kept in the history array the_fish::fish_skel::history::energy_balance_curr. More... | |
| elemental real(srp) function | fish_energy_branchial_step (this, temperature, is_oxygen) |
| Calculate urinal (UE) and branchial energy (ZE) as a proportion of the SMR. More... | |
| elemental real(srp) function | nitrogen_mass (mol, is_ammonia) |
| Calculate the mass of ammonia, mol to g. More... | |
| elemental real(srp) function | branchial_nitrogen_to_energy (excreted_nitrogen, is_kg) |
| Calculate the branchal and urinary energy loss from branchial and urinary nitrogen loss. See Bureau, D.P., Kaushik, S.J., Cho, C.Y. (2002) Bioenergetics. In Hardy, R.W., Halver, J.E., eds. Fish Nutrition, 3 ed Academic Press, San Diego, CA, USA (pp 1–59): More... | |
| elemental real(srp) function | fish_energy_consumption_step (this, temperature, sda_off) |
| Calculate total energy, kJ consumption of the fish depending on the temperature E = SMR + AMR + (UE+ZE) + SDA Public interface: the_fish::uptake_energy(). More... | |
| elemental real(srp) function | fish_oxygen_consumption_step (this, temperature, is_mass, sda_off) |
| Calculate total oxygen consumption (l) of the fish depending on the temperature O2 = O2_SMR + O2_AMR + O2_{UE+ZE} O2_SDA. More... | |
| elemental real(srp) function | fish_energy_sda_factor (this, max_ap, max_sda, is_limit) |
Calculate the relative SDA (specific dynamic action) factor for SMR unit of energy uptake. SDA depends on the absorption rate up th the maxmum cap value max_sda that is achieved at the absorption rate max_ap More... | |
| elemental subroutine | fish_grow_increment_energy (this) |
| Grow the fish body mass: increment mass based on energy. More... | |
| elemental real(srp) function | fish_midgut_total_mass_evacuated_step (this, max_time) |
| Calculate the increment of the total mass of food evacuated from the midgut after full absorption and evacuation delay (see commondata:global_maximum_duration_midgut_min) that occurs at one time step. More... | |
| elemental integer function | fish_midgut_n_evacuated_step (this, max_time) |
| Calculate the number of the total mass of food evacuated from the midgut after full absorption and evacuation delay (see commondata:global_maximum_duration_midgut_min) that occurs at one time step. More... | |
| elemental subroutine | fish_midgut_history_update (this) |
| Update the history stack arrays for the fish, ::midgut class. More... | |
| elemental integer function | midgut_absorp_history_last_past_idx (this) |
| Determine the index of the latest absorption item in the history stack array. If the whole array is commondata::missing, this means the history stack is empty, then the function returns zero. Note that in most cases the function should return commondata::history_size as the history stack is filled with previous values. More... | |
| elemental real(srp) function | fish_midgut_total_mass_food (this, tolerance) |
| Calculate the total mass of food in the fish midgut. More... | |
| elemental real(srp) function | fish_appetite_factor_midgut (this, fcapacity) |
| Calculate the fish appetite factor based on the relative midgut fullness. Appetite factor is equivalent to the probability of consuming any food item. More... | |
| elemental real(srp) function | fish_midgut_absorption_rate (this) |
| Calculate instantaneous absorption rate based on the current absorption data and previous absorption saved in Output Arrays. More... | |
| elemental real(srp) function | fish_midgut_absorption_factor (this) |
| Appetite factor multiplier based on the total absorption rate. More... | |
| elemental subroutine | fish_midgut_shift_items_after_new_ingest (this, food_item_pass) |
| Shift food items in the midgut after a new food item is ingested and therefore food items are shifted in stomach adding the new item. More... | |
| elemental real(srp) function | appetite_func (mass_relative, A, R) |
| Computational backend for the appetite function. Appetite is calculated as. More... | |
| elemental real(srp) function | fish_appetite_probability_ingestion (this, time_step) |
| Calculate the overall appetite of the fish, which defines the probability to ingest any food item. More... | |
| pure real(srp) function | stress_suppress_factor_s (time_step_incr, is_force) |
Calculate suppression factor for appetite caused by stress intervention at time time: generic interface is ::stress_factor(). This is the scalar version, vector -based version is stress_suppress_factor_v(). More... | |
| pure real(srp) function, dimension(:), allocatable | stress_suppress_factor_v (time_step_incr, is_force) |
| Vector-based version of the stress-based suppression function ::stress_factor(). There is a scalar version of the same function stress_suppress_factor_s. More... | |
| pure real(srp) function | stress_suppress (time_step) |
| Calculate real stress suppression factor for a specific time step given the configured stress intervention pattern. This is a high-level interface to ::stress_factor(). More... | |
| elemental real(srp) function | fish_body_mass_current_get (this) |
| Get the current body mass of the fish. Note that this is the shortcut to obtain the last value from the body mass history array the_fish::fish_skel::history::body_mass_current. More... | |
| elemental real(srp) function | fish_appetite_factor_energy_balance (this) |
| Calculate appetite factor that depends on the energy balance. More... | |
| impure subroutine | fish_decide_eat_food_item (this, food_item_in, have_eaten) |
| This procedure determines if the fish decides to consume the food item and if yes, ingests it using the ::do_ingest() method. @important This version accepts raw food item. Generic interface linking both raw and pointer versions is ambiguous. More... | |
| impure subroutine | fish_decide_eat_food_item_ptr (this, food_item_in, have_eaten) |
| This procedure determines if the fish decides to consume the food item and if yes, ingests it using the ::do_ingest() method. @important This version accepts pointer to food item. Generic interface linking both raw and pointer versions is ambiguous. More... | |
| elemental real(srp) function | fish_activity_baseline_component (this, time_step) |
| Calculate the baseline activity of the fish. More... | |
| elemental real(srp) function | fish_activity_appetite_factor (this) |
| Calculate activity factor linked to appetite. Appetite factor has a linear relationship with the fish appetite !> | |
| elemental real(srp) function | stomach_adjust (target_emptying_time) |
| Calculate the adjustment factor for the transformation of the stomach transport pattern Time scale commondata::global_transport_pattern_t for a different temperature or fish size. More... | |
| elemental integer function | stomach_emptying_time (fish_mass, temperature) |
| Calculate the stomach emptying time for a fish with specific body mass at a specific temperature. Calculation is based on spline interpolation of experimental data. More... | |
| elemental integer function | fish_stomach_emptying_time (this, temperature) |
| Object-oriented frontend for stomach_emptying_time(): Calculate the stomach emptying time for a fish with specific body mass at a specific temperature. Calculation is based on spline interpolation of experimental data. More... | |
| impure type(stomach_emptying_pattern) function | get_stomach_emptying_matrix_csv (csv_file) |
| Get the stomach emptying time base matrix from a CSV input data file. More... | |
Variables | |
| integer, parameter, public | history_size = 20 |
| The size of the history that saves the fish data back in time. More... | |
Detailed Description
This module defines the fish and its components. The model is discrete, time is based on time steps, in s.
Function/Subroutine Documentation
◆ stomach_vol()
| elemental real(srp) function the_fish::stomach_vol | ( | real(srp), intent(in) | mass | ) |
Calculate the stomach volume (ml) based on the fish body mass (g) using Salgado, A., Salgado, I., & Valdebenito, I. (2018). A direct and straightforward method for measurement real maximum fish stomach volume to improve aquaculture feeding research. Latin American Journal of Aquatic Research, 46(5), 880–889. https://doi.org/10.3856/vol46-issue5-fulltext-3.
- Parameters
-
[in] mass fish body mass
Definition at line 308 of file m_fish.f90.
◆ fish_init_new_zero()
| elemental subroutine the_fish::fish_init_new_zero | ( | class(fish_skel), intent(inout) | this, |
| integer, intent(in), optional | id_num | ||
| ) |
Initialise an empty ::fish object to an empty zero state.
- Parameters
-
[in] id_num Optional ID number of the fish, default is commondata::unknown
Optional appetite factor level of the fish. Default is commondata::missing
Definition at line 318 of file m_fish.f90.
◆ fish_smr_temp_bodymass_evan1990_scalar()
| pure real(srp) function the_fish::fish_smr_temp_bodymass_evan1990_scalar | ( | class(fish_skel), intent(in) | this, |
| real(srp), intent(in) | temperature, | ||
| logical, intent(in), optional | is_per_hour, | ||
| integer(long), intent(in), optional | time_step | ||
| ) |
SMR calculation from Evans 1992 data, scalar version.
- Parameters
-
[in] temperature Ambient temperature ºC [in] is_per_hour Optional flag if calculation is done per hour (default FALSE, meaning per one time step, second)
- Returns
- Standard metabolic rate (SMR) in mg O2 / kg per unit time (s or h)
Definition at line 346 of file m_fish.f90.
◆ fish_smr_temp_bodymass_evan1990_value()
| pure real(srp) function the_fish::fish_smr_temp_bodymass_evan1990_value | ( | real(srp), intent(in) | body_mass, |
| real(srp), intent(in) | temperature, | ||
| logical, intent(in), optional | is_per_hour, | ||
| integer(long), intent(in), optional | time_step | ||
| ) |
SMR calculation from Evans 1992 data, non-object scalar version.
- Parameters
-
[in] body_mass Body mass of the fish, kg [in] temperature Ambient temperature ºC [in] is_per_hour Optional flag if calculation is done per hour (default FALSE, per second)
- Returns
- Standard metabolic rate (SMR) in mg O2 / kg per unit time (s or h)
Definition at line 381 of file m_fish.f90.
◆ amr_swim_basic()
| elemental real(srp) function the_fish::amr_swim_basic | ( | real(srp), intent(in) | fish_mass, |
| real(srp), intent(in) | swimming_speed, | ||
| integer, intent(in), optional | time_unit | ||
| ) |
Calculate the active metabolic rate of fish in units of oxygen consumption per hour.
![]()
See Ohlberger, J., Staaks, G., Van Dijk, P. L. M., & Hölker, F. (2005). Modelling energetic costs of fish swimming. Journal of Experimental Zoology Part A: Comparative Experimental Biology, 303, 657–664.
- Parameters
-
[in] fish_mass Fish mass, g [in] swimming_speed Fish swimming speed cm/s [in] time_unit Optional time unit, e.g. commondata::minute or commondata::hour The default value is s 1/60 of commondata:::minute
- Returns
- Active metabolic rate, mg O2 per
time_unit(equation default commondata::hour)
Definition at line 431 of file m_fish.f90.
◆ fish_amr_locomotion_energy_cost()
| elemental real(srp) function the_fish::fish_amr_locomotion_energy_cost | ( | class(fish), intent(in) | this, |
| real(srp), intent(in), optional | temperature, | ||
| logical, intent(in), optional | is_exclude_smr, | ||
| logical, intent(in), optional | is_per_hour | ||
| ) |
Calculate the active metabolic rate. This can be based on amr_swim_basic() or def_salmon::salmon_oxygen_consumption().
- Parameters
-
[in] temperature Ambient temperature, C [in] is_exclude_smr Opional flag to force exclude SMR (if SMR is included into the equation making this function essentially SMR + AMR function). Note that SMR and AMR are added in the_fish::fish::energy_update(). The default value is FALSE. [in] is_per_hour Optional flag if calculation is done per hour (default FALSE, meaning per one time step, second)
- Returns
- Active metabolic rate (AMR), mg O2 / s
Definition at line 472 of file m_fish.f90.
◆ fish_whole_body_energy_trout()
| elemental real(srp) function the_fish::fish_whole_body_energy_trout | ( | class(fish_skel), intent(in) | this | ) |
Calculate the whole body energy content.
- Returns
- Energy content of the body, kJ per whole fish
![]()
Definition at line 525 of file m_fish.f90.
◆ energy2mass()
| elemental real(srp) function the_fish::energy2mass | ( | real(srp), intent(in) | body_energy | ) |
Calculate the body mass from whole body energy.
- Note
- Note that this procedure is the reverse of the_fish::fish_whole_body_energy_trout().
- Parameters
-
[in] body_energy Whole body energy of the fish, kJ
- Returns
- Body mass, g
![]()
Definition at line 546 of file m_fish.f90.
◆ food_item_eaten_set_zero_init_state()
| elemental subroutine the_fish::food_item_eaten_set_zero_init_state | ( | class(food_item_eaten), intent(inout) | this, |
| real(srp), intent(in), optional | mass, | ||
| real(srp), intent(in), optional | start_mass, | ||
| integer, intent(in), optional | delay | ||
| ) |
Initialise an empty food item as ingested by the fish. Cleanup and set zero state.
- Warning
- This procedure is by default used as a destructor, it sets the food item an undefined state (missing, unknown).
- Parameters
-
[in] mass Optional mass of the food item, default is set to commondata::global_food_item_mass [in] start_mass Optional starting reference mass of the food item.
- Note
- Not used here
- Parameters
-
[in] delay Optional ingestion delay, default is set from the parameter commondata::global_ingestion_delay
Definition at line 565 of file m_fish.f90.
◆ food_item_eaten_mg_set_zero_init_state()
| elemental subroutine the_fish::food_item_eaten_mg_set_zero_init_state | ( | class(food_item_eaten_mg), intent(inout) | this, |
| real(srp), intent(in), optional | mass, | ||
| real(srp), intent(in), optional | start_mass, | ||
| integer, intent(in), optional | delay | ||
| ) |
Initialise an empty food item as ingested by the fish. Cleanup and set zero state.
- Warning
- This procedure is by default used as a destructor, it sets the food item an undefined state (missing, unknown).
- Parameters
-
[in] mass Optional mass of the food item, default is set to commondata::global_food_item_mass [in] start_mass Optional starting reference mass of the food item. [in] delay Optional ingestion delay, default is set from the parameter commondata::global_ingestion_delay
Definition at line 600 of file m_fish.f90.
◆ food_item_eaten_eat_init_at_ingest()
| elemental subroutine the_fish::food_item_eaten_eat_init_at_ingest | ( | class(food_item_eaten), intent(inout) | this, |
| class(food_item), intent(in) | food_item_in, | ||
| integer, intent(in), optional | ingest_delay | ||
| ) |
Initialise a single food item as it is ingested by the fish. Note that the ingested food item is copied from the external food item environ::food_item that comes as the input argument.
- Parameters
-
[in] food_item_in The food item object (class) being ingested by the fish. [in] ingest_delay Optional ingestion delay, if not provided the default value is commondata::global_ingestion_delay.
- Note
- Ingestion delay is assumed a property of the fish gastrointestinal system rather than the food item.
- copy all data components from the input argument
food_item_intothis. Note that class assignment operator overloading is used for=assignment with backend defined in environ::food_item_copy_assign_to(). - The time counter of the food item within the gastrointestinal tract of the fish is set to zero.
Definition at line 651 of file m_fish.f90.
◆ food_item_eaten_update_age_in_stomach()
| elemental subroutine the_fish::food_item_eaten_update_age_in_stomach | ( | class(food_item_eaten), intent(inout) | this, |
| integer, intent(in), optional | t_increment | ||
| ) |
This procedure updates the time the food item spent in the stomach of the fish.
- Parameters
-
[in] t_increment Optional time step increment, default value is 1.
Definition at line 685 of file m_fish.f90.
◆ food_item_eaten_check_destruct()
| elemental subroutine the_fish::food_item_eaten_check_destruct | ( | class(food_item_eaten), intent(inout) | this, |
| logical, intent(out), optional | is_destructed, | ||
| real(srp), intent(in), optional | tolerance, | ||
| integer, intent(in), optional | max_time | ||
| ) |
Check if the food item mass shrunk to near-zero and destruct it.
- Parameters
-
[out] is_destructed Optional logical indicator that is set to TRUE if the food item was actually destructed. [in] tolerance Optional numerical tolerance, the smallest value of the food item mass that counts as non-zero. Default is set by commondata::min_vol_toler. [in] max_time Maximum time parameter, not used here, for compatibility with the respective midgut method
Definition at line 703 of file m_fish.f90.
◆ food_item_eaten_mg_check_destruct()
| elemental subroutine the_fish::food_item_eaten_mg_check_destruct | ( | class(food_item_eaten_mg), intent(inout) | this, |
| logical, intent(out), optional | is_destructed, | ||
| real(srp), intent(in), optional | tolerance, | ||
| integer, intent(in), optional | max_time | ||
| ) |
Check if the food item has been processed for more than commondata::global_maximum_duration_midgut s time in the midgut.
- Parameters
-
[out] is_destructed Optional logical indicator that is set to TRUE if the food item was actually destructed. [in] tolerance Optional numerical tolerance parameter. Note that it is not used here but needed for compatibility with the food_item_eaten::zero() method implemented for the base type the_fish::food_item_eaten. [in] max_time Maximum time processed in the midgut, by default all food items that have been in the process of absorption for more than this time will destructed from midgut and evacuated.
The food item is destructed with the food_item_eaten_mg::zero() function.
Definition at line 737 of file m_fish.f90.
◆ food_item_eaten_mg_exceed_max_is_to_destruct()
| elemental logical function the_fish::food_item_eaten_mg_exceed_max_is_to_destruct | ( | class(food_item_eaten_mg), intent(in) | this, |
| integer, intent(in), optional | max_time | ||
| ) |
Check if the food item in the midgut has spent for more than the maximum time in the midgut after full absorption and should be destructed and evacuated (TRUE).
- Parameters
-
[in] max_time Maximum time processed in the midgut, by default all food items that have been in the process of absorption for more than this time will destructed from midgut and evacuated. Default value is based on commondata::global_maximum_duration_midgut_min.
Definition at line 775 of file m_fish.f90.
◆ food_item_eaten_is_absorbed_full()
| elemental logical function the_fish::food_item_eaten_is_absorbed_full | ( | class(food_item_eaten_mg), intent(in) | this, |
| real(srp), intent(in), optional | max_absorb | ||
| ) |
Check if the food item has been absorbed to the maximum possible mass.
Definition at line 803 of file m_fish.f90.
◆ st_food_item_mass()
| elemental real(srp) function the_fish::st_food_item_mass | ( | integer, intent(in) | time_in_process, |
| real(srp), intent(in), optional | dry_mass, | ||
| integer, intent(in), optional | ingestion_delay, | ||
| real(srp), intent(in), optional | water_uptake, | ||
| real(srp), intent(in), optional | rescale | ||
| ) |
This function defines the mass of a single food item as it passes through the fish stomach.
- Parameters
-
[in] time_in_process The time since the food item has been ingested by the fish [in] dry_mass Optional dry mass of the food item, if absent the default value commondata::global_food_item_mass is used. [in] ingestion_delay Ingestion delay: the time duration from ingestion of the food item until it starts transport into midgut. Water uptake occurs during the ingestion delay. Default is commondata::global_ingestion_delay. [in] water_uptake The relative mass of water added to the food item after ingestion during the ingestion delay. Default is commondata::global_water_uptake. [in] rescale Optional time rescaling parameter for the stomach transport array
- Returns
- Returns the mass of the food item.
Implementation notes
In stomach, the feed that is ingested is passed through two stages:
- ingestion delay (commondata::global_ingestion_delay) when water uptake occurs and the mass of the food item increases:
- passage to midgut where the mass of the food item decreases.
The food item is subjected to water uptake. The pattern of water uptake over time, the maximum the_fish::food_item_eaten::ingestion_delay seconds, is defined by a commondata::logistic() equation:
![]()
where ![]() is the dry mass of the food item, as defined by the parameter commondata::global_food_item_mass,
is the dry mass of the food item, as defined by the parameter commondata::global_food_item_mass, ![]() is the asymptotic maximum mass of the food item after water intake,
is the asymptotic maximum mass of the food item after water intake, ![]() and
and ![]() are logistic function parameters (see commondata::logistic()).
are logistic function parameters (see commondata::logistic()).
- The asymptotic maximum mass of the food item after water uptake
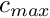 is defined by
is defined by
where![\[ c_{max} = c_{0} + c_{0} \cdot u \]](form_20.png)
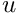 is the water uptake parameter commondata::global_water_uptake.
is the water uptake parameter commondata::global_water_uptake.
The value of the function is calculated as follows.
- If the time the food item spend in the stomach is smaller than the ingestion delay (commondata::global_ingestion_delay), its mass increases through water uptake up to the
max_volvalue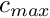 .
.
If the time the food item spent in stomach is longer than the ingestion delay, its mass decreases (from the maximum mass after water intake) as determined by the interpolation function that is determined by the grid:
- abscissa: commondata::global_transport_pattern_t;
- ordinate: commondata::global_transport_pattern_r.
given the time after water intake completed (ingestion delay commondata::global_ingestion_delay).
Definition at line 835 of file m_fish.f90.
◆ fish_stomach_init()
| elemental subroutine the_fish::fish_stomach_init | ( | class(stomach), intent(inout) | this, |
| real(srp), intent(in), optional | mass | ||
| ) |
Initialise an empty fish stomach object.
- Warning
- Note that the fish itself is not initialised here, it should be done with the ::fish::new() method.
Note that at the init, all food items in the stomach are destructed with the ::food_item_eaten::zero() method.
Definition at line 948 of file m_fish.f90.
◆ fish_stomach_update_step()
| impure subroutine the_fish::fish_stomach_update_step | ( | class(stomach), intent(inout) | this | ) |
Update one time step of the fish stomach life cycle.
Implementation notes
First, the time counter for each of the existing (non-zero) n_food_items_stomach food item (time_in_process) is updated, by default incremented by 1.
Second, the mass of each food item in stomach is recalculated according to its individual time_in_process counter.
Check if the food items shrunk below the minimum size and should then disappear (be destructed). The is_destruct boolean indicators show how many food items should be destructed
In the debug mode, also check and fix counter for negative values.
Definition at line 968 of file m_fish.f90.
◆ fish_ingest_food_item()
| elemental subroutine the_fish::fish_ingest_food_item | ( | class(fish), intent(inout) | this, |
| class(food_item), intent(in) | food_item_ingested | ||
| ) |
This procedure shows what happens when the fish eats a single food item.
- Parameters
-
[in] food_item_ingested An environ::food_item class object that represents the food item being ingested.
- Individual totals are updated: N of food items ingested and the total mass of food ingested.
- The newly ingested food item object is initialised, data structure is copied from the input argument food_item object. Ingestion delay is set default.
- The food item is then added to the stack array of the fish stomach. It takes the first place shifting all other food items in sequence.
- The food items counter is incremented.
- Finally, whenever the food is eaten, initialise it in the midgut.
Definition at line 1025 of file m_fish.f90.
◆ fish_stomach_total_mass_food()
| elemental real(srp) function the_fish::fish_stomach_total_mass_food | ( | class(stomach), intent(in) | this, |
| real(srp), intent(in), optional | tolerance | ||
| ) |
Calculate the total mass of food in the fish stomach.
- Parameters
-
[in] tolerance Optional numerical tolerance, the smallest value of the food item mass that counts as non-zero. Default is set by commondata::min_vol_toler.
- Returns
- Total mass of food in the stomach.
The total value is calculated as an array sum to the total number of food items being kept in the stomach (thisn_food_items_stomach).
Definition at line 1064 of file m_fish.f90.
◆ fish_appetite_factor_stomach()
| elemental real(srp) function the_fish::fish_appetite_factor_stomach | ( | class(stomach), intent(in) | this, |
| real(srp), intent(in), optional | fcapacity | ||
| ) |
Calculate the fish appetite factor based on the relative stomach fullness. Appetite factor is equivalent to the probability of consuming any food item.
Implementation notes
The appetite factor ranges between (0 and 1) and is based on the relative filling capacity. Here relative filling capacity ![]() is calculated as the ratio of the total mass of food in the stomach
is calculated as the ratio of the total mass of food in the stomach ![]() to the maximum filling capacity
to the maximum filling capacity ![]()
![]()
Given the relative filling capacity ![]() , appetite factor that ranges from 0 to 1 is calculated as logistic function appetite_func().
, appetite factor that ranges from 0 to 1 is calculated as logistic function appetite_func().
The ![]() and
and ![]() logistic parameters are defined by the commondata::global_appetite_logist_a and commondata::global_appetite_logist_r parameters.
logistic parameters are defined by the commondata::global_appetite_logist_a and commondata::global_appetite_logist_r parameters.
gnuplot code:
set xrange [0:1]
plot 1 - 1/(1+500000*exp(-20*x))
Definition at line 1096 of file m_fish.f90.
◆ fish_midgut_init()
| elemental subroutine the_fish::fish_midgut_init | ( | class(midgut), intent(inout) | this, |
| real(srp), intent(in), optional | mass | ||
| ) |
Initialise an empty fish midgut object.
- Warning
- Note that the fish itself is not initialised here, it should be done with the ::fish::new() method.
Note that at the init, all food items in the stomach are destructed with the ::food_item_eaten::zero() method.
Definition at line 1142 of file m_fish.f90.
◆ fish_midgut_update_from_stomach()
| impure subroutine the_fish::fish_midgut_update_from_stomach | ( | class(midgut), intent(inout) | this, |
| real(srp), intent(in), optional | dry_mass, | ||
| integer, intent(in), optional | t_increment | ||
| ) |
This procedure processes the food item in the midgut of the fish.
- Parameters
-
[in] dry_mass Optional dry mass of the food item, if absent the default value commondata::global_food_item_mass is used. [in] t_increment Optional time step increment, default value is 1.
Implementation notes
Transfer to midgut
Each food item smoothly transfers from the stomach to the midgut, which is implemented as removing a small proportion if its mass with simultaneous addition of the same mass to the linked midgut data component.
First, for each valid food item in the stomach, calculate the difference between mass of the food item in stomach at the previous time step t-1 and the current step t: ![]() (
(delta_vol).
![]()
where ![]() is the mass of the i-th food item in the stomach. This difference is equal to the mass increment that transfers to the midgut.
is the mass of the i-th food item in the stomach. This difference is equal to the mass increment that transfers to the midgut.
This increment is set to zero for all food items that stayed in the stomach for less than the ingestion_delay duration (this period is water uptake, no transfer, see stomach::st_step()).
The increment ![]() (
(delta_vol) is here also set to zero for all food items that have transferred fully from the stomach to midgut.
Second, the mass of each food item already in the midgut is augmented by the respective increment ![]() .
.
![]()
The age of each food item (time_in_process data component) in the midgut is also incremented by one (i.e. discretization period, s).
The timer for the duration spent in the midgut is also incremented.
Absorption
The absorption processis this: certain proportion of the mass of each food item in the midgut is subtracted following the equation:
![]()
where ![]() and
and ![]() are the Michaelis-Menten equation parameters: the maximum rate and the K constant (see commondata::michaelis_menten() for more details). Here
are the Michaelis-Menten equation parameters: the maximum rate and the K constant (see commondata::michaelis_menten() for more details). Here ![]() is the total mass of food in the midgut.
is the total mass of food in the midgut.
Only the food items that stay in the midgut for more than the duration defined by commondata::global_digestion_delay_min are subject to absorption. Also, there is a maximum limit on the absorption set by the commondata::global_absorption_ratio model parameter. This limit is checked in the food_item_eaten_mg::is_absorbed_full() method.
- Note
- Note that each food item in the midgut is subjected to Michaelis-Menten absorption depending on the total mass of food in the midgut
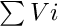 .
.
Finally, the food_item_eaten_mg::check_disappear() method is called for each food item in the midgut that has fully transferred from the stomach. This checks if the item has been processed in the midgut or more than commondata::global_maximum_duration_midgut_min. Those food items are deleted from the midgut and are excreted.
The time counter for the time since full absorption (time_absorbed data component) also updates (incremented by 1) for all food items that are fully absorbed.
Update the individual totals for the mass of food evacuated.
Check if the food items must be evacuated from the midgut, disappear
The count of the food items in the midgut is updated after reduction at the previous step.
Definition at line 1162 of file m_fish.f90.
◆ fish_energy_balance_update()
| elemental subroutine the_fish::fish_energy_balance_update | ( | class(fish), intent(inout) | this, |
| real(srp), intent(in), optional | temperature, | ||
| real(srp), intent(in), optional | start_value | ||
| ) |
Calculate and update the current energy balance of the fish. Note that the energy balance is kept in the history array the_fish::fish_skel::history::energy_balance_curr.
- Parameters
-
[in] temperature Optional current ambient temperature, if the parameter absent, use the global default commondata::global_temperature. [in] start_value Optional start value of the energy balance, if the parameter is absent, the nefault value oz zero is used
Implementation notes
![]()
where ![]() is the energy balance of the fish at time
is the energy balance of the fish at time ![]() (history array: the_fish::history_array_fish::energy_balance_curr),
(history array: the_fish::history_array_fish::energy_balance_curr), ![]() is the energy that came from increment in the food absorption
is the energy that came from increment in the food absorption ![]() ,
, ![]() is the energetic equivalent of the standard metabolic rate based on the_fish::fish_skel::smr(), and
is the energetic equivalent of the standard metabolic rate based on the_fish::fish_skel::smr(), and ![]() is the energetic equivalent of the active metabolic rate bsed on the_fish::fish_skel::amr().
is the energetic equivalent of the active metabolic rate bsed on the_fish::fish_skel::amr().
Definition at line 1435 of file m_fish.f90.
◆ fish_energy_branchial_step()
| elemental real(srp) function the_fish::fish_energy_branchial_step | ( | class(fish), intent(in) | this, |
| real(srp), intent(in), optional | temperature, | ||
| logical, intent(in), optional | is_oxygen | ||
| ) |
Calculate urinal (UE) and branchial energy (ZE) as a proportion of the SMR.
- Parameters
-
[in] temperature Optional current ambient temperature, if the parameter absent, use the global default commondata::global_temperature. [in] is_oxygen Optional logical flag that defines that oxygen consumption (mass, g) rather than energy is output, default is FALSE (energy is calculated)
- Note
- Note that oxygen consumption in case of
is_oxygen=TRUEis output as mass in g, NOT volume.
Definition at line 1501 of file m_fish.f90.
◆ nitrogen_mass()
| elemental real(srp) function the_fish::nitrogen_mass | ( | real(srp), intent(in) | mol, |
| logical, intent(in), optional | is_ammonia | ||
| ) |
Calculate the mass of ammonia, mol to g.
- Parameters
-
[in] mol Amount in mol [in] is_ammonia Optional flag indicating that
Definition at line 1553 of file m_fish.f90.
◆ branchial_nitrogen_to_energy()
| elemental real(srp) function the_fish::branchial_nitrogen_to_energy | ( | real(srp), intent(in) | excreted_nitrogen, |
| logical, intent(in), optional | is_kg | ||
| ) |
Calculate the branchal and urinary energy loss from branchial and urinary nitrogen loss. See Bureau, D.P., Kaushik, S.J., Cho, C.Y. (2002) Bioenergetics. In Hardy, R.W., Halver, J.E., eds. Fish Nutrition, 3 ed Academic Press, San Diego, CA, USA (pp 1–59):
<<Because most nitrogen losses are as ammonia, and the difference in the amount of energy loss per gram of nitrogen between ammonia and urea is small, it has been proposed that the loss of1gofnitrogen by fish under normal conditions can be equated to an energy loss of 24.9 kJ.>> (p.21)
see also Saravanan, S., Schrama, J. W., Figueiredo-Silva, A. C., Kaushik, S. J., Verreth, J. A. J., & Geurden, I. (2012). Constraints on Energy Intake in Fish: The Link between Diet Composition, Energy Metabolism, and Energy Intake in Rainbow Trout. PLoS ONE, 7(4), e34743. https://doi.org/10.1371/journal.pone.0034743
- Parameters
-
[in] excreted_nitrogen Branchial and urinary nitrogen loss, g [in] is_kg Optional flag indicating that excreted nitrogen loss is given in kg
- Returns
- Energy loss kJ
Energy equivalent kJ of 1 g excreted nitrogen NH3-N
Definition at line 1595 of file m_fish.f90.
◆ fish_energy_consumption_step()
| elemental real(srp) function the_fish::fish_energy_consumption_step | ( | class(fish), intent(in) | this, |
| real(srp), intent(in), optional | temperature, | ||
| logical, intent(in), optional | sda_off | ||
| ) |
Calculate total energy, kJ consumption of the fish depending on the temperature E = SMR + AMR + (UE+ZE) + SDA Public interface: the_fish::uptake_energy().
- Note
- This function should be adapted together with the oxygen consumption code fish_oxygen_consumption_step().
- Parameters
-
[in] sda_off Optional logical switch that sets SDA component OFF, by default FALSE, intended for use mainly for debugging and testing.
Definition at line 1625 of file m_fish.f90.
◆ fish_oxygen_consumption_step()
| elemental real(srp) function the_fish::fish_oxygen_consumption_step | ( | class(fish), intent(in) | this, |
| real(srp), intent(in), optional | temperature, | ||
| logical, intent(in), optional | is_mass, | ||
| logical, intent(in), optional | sda_off | ||
| ) |
Calculate total oxygen consumption (l) of the fish depending on the temperature O2 = O2_SMR + O2_AMR + O2_{UE+ZE} O2_SDA.
- Note
- This function should be adapted together with the energy consumption code fish_energy_consumption_step().
- Parameters
-
[in] is_mass Optional logical switch, if TRUE, determining that oxygen consumption is calculated as mass (g), the default is volume (l) [in] sda_off Optional logical switch that sets SDA component OFF, by default FALSE, intended for use mainly for debugging and testing.
Definition at line 1667 of file m_fish.f90.
◆ fish_energy_sda_factor()
| elemental real(srp) function the_fish::fish_energy_sda_factor | ( | class(fish), intent(in) | this, |
| real(srp), intent(in) | max_ap, | ||
| real(srp), intent(in) | max_sda, | ||
| logical, intent(in), optional | is_limit | ||
| ) |
Calculate the relative SDA (specific dynamic action) factor for SMR unit of energy uptake. SDA depends on the absorption rate up th the maxmum cap value max_sda that is achieved at the absorption rate max_ap
The relative SDA factor is based on a linear relationship defined by the maximum absorption rate max_ap and max SDA factor max_sda.
max_sda + * 2,0 :: SDA = 2.0 x SMR
| * . y = k x + b
| * . ? =0
| * .
0 +-------+
0 max_ap = 0.0007 - Parameters
-
[in] max_ap Maximum (absolute) absorption rate when the maximum value of SDA ( max_sda) is achieved[in] max_sda Maximum value of SDA achieved at the maximum absorption rate ( max_ap)
Definition at line 1746 of file m_fish.f90.
◆ fish_grow_increment_energy()
| elemental subroutine the_fish::fish_grow_increment_energy | ( | class(fish), intent(inout) | this | ) |
Grow the fish body mass: increment mass based on energy.
Implementation details
The current body mass is incremented as ![]() by the mass equivalent
by the mass equivalent ![]() of the energy increment for the time step calculated using the_fish::energy2mass().
of the energy increment for the time step calculated using the_fish::energy2mass().
Definition at line 1774 of file m_fish.f90.
◆ fish_midgut_total_mass_evacuated_step()
| elemental real(srp) function the_fish::fish_midgut_total_mass_evacuated_step | ( | class(midgut), intent(in) | this, |
| integer, intent(in), optional | max_time | ||
| ) |
Calculate the increment of the total mass of food evacuated from the midgut after full absorption and evacuation delay (see commondata:global_maximum_duration_midgut_min) that occurs at one time step.
- Warning
- Note that this method should be called before the_fish::food_item_eaten_mg::check_disappear() so that all food itemsthat should be evacuated have not yet been evacuated.
- Parameters
-
[in] max_time Maximum time processed in the midgut, by default all food items that have been in the process of absorption for more than this time will destructed from midgut and evacuated. Default value is based on commondata::global_maximum_duration_midgut_min.
Definition at line 1811 of file m_fish.f90.
◆ fish_midgut_n_evacuated_step()
| elemental integer function the_fish::fish_midgut_n_evacuated_step | ( | class(midgut), intent(in) | this, |
| integer, intent(in), optional | max_time | ||
| ) |
Calculate the number of the total mass of food evacuated from the midgut after full absorption and evacuation delay (see commondata:global_maximum_duration_midgut_min) that occurs at one time step.
- Warning
- Note that this method should be called before the_fish::food_item_eaten_mg::check_disappear() so that all food itemsthat should be evacuated have not yet been evacuated.
- Parameters
-
[in] max_time Maximum time processed in the midgut, by default all food items that have been in the process of absorption for more than this time will destructed from midgut and evacuated. Default value is based on commondata::global_maximum_duration_midgut_min.
Definition at line 1854 of file m_fish.f90.
◆ fish_midgut_history_update()
| elemental subroutine the_fish::fish_midgut_history_update | ( | class(midgut), intent(inout) | this | ) |
Update the history stack arrays for the fish, ::midgut class.
Definition at line 1880 of file m_fish.f90.
◆ midgut_absorp_history_last_past_idx()
| elemental integer function the_fish::midgut_absorp_history_last_past_idx | ( | class(midgut), intent(in) | this | ) |
Determine the index of the latest absorption item in the history stack array. If the whole array is commondata::missing, this means the history stack is empty, then the function returns zero. Note that in most cases the function should return commondata::history_size as the history stack is filled with previous values.
Definition at line 1927 of file m_fish.f90.
◆ fish_midgut_total_mass_food()
| elemental real(srp) function the_fish::fish_midgut_total_mass_food | ( | class(midgut), intent(in) | this, |
| real(srp), intent(in), optional | tolerance | ||
| ) |
Calculate the total mass of food in the fish midgut.
- Parameters
-
[in] tolerance Optional numerical tolerance, the smallest value of the food item mass that counts as non-zero. Default is set by commondata::min_vol_toler.
- Returns
- Total mass of food in the midgut.
The total value is calculated as an array sum to the total number of food items being kept in the stomach (thisn_food_items_stomach).
Definition at line 1941 of file m_fish.f90.
◆ fish_appetite_factor_midgut()
| elemental real(srp) function the_fish::fish_appetite_factor_midgut | ( | class(midgut), intent(in) | this, |
| real(srp), intent(in), optional | fcapacity | ||
| ) |
Calculate the fish appetite factor based on the relative midgut fullness. Appetite factor is equivalent to the probability of consuming any food item.
- Note
- Note that the parameters of the appetite function are identical for both the stomach and midgut appetite factors:
- stomach::appetite_stomach
- midgut::
Implementation notes
The appetite factor ranges between (0 and 1) and is based on the relative filling capacity. Here relative filling capacity ![]() is calculated as the ratio of the total mass of food in the midgut
is calculated as the ratio of the total mass of food in the midgut ![]() to the maximum filling capacity
to the maximum filling capacity ![]()
![]()
Correction factor is introduced to take account of the fact that the capacity of the midgut is bigger than stomach but basically the same amount of food is coming from stomach (in case of one meal).
Given the relative filling capacity ![]() , appetite factor that ranges from 0 to 1 is calculated as logistic function appetite_func().
, appetite factor that ranges from 0 to 1 is calculated as logistic function appetite_func().
The ![]() and
and ![]() logistic parameters are defined by the commondata::global_appetite_logist_a and commondata::global_appetite_logist_r parameters.
logistic parameters are defined by the commondata::global_appetite_logist_a and commondata::global_appetite_logist_r parameters.
gnuplot code:
set xrange [0:1]
plot 1 - 1/(1+500000*exp(-20*x))
Definition at line 1979 of file m_fish.f90.
◆ fish_midgut_absorption_rate()
| elemental real(srp) function the_fish::fish_midgut_absorption_rate | ( | class(midgut), intent(in) | this | ) |
Calculate instantaneous absorption rate based on the current absorption data and previous absorption saved in Output Arrays.
Definition at line 2029 of file m_fish.f90.
◆ fish_midgut_absorption_factor()
| elemental real(srp) function the_fish::fish_midgut_absorption_factor | ( | class(midgut), intent(in) | this | ) |
Appetite factor multiplier based on the total absorption rate.
Definition at line 2047 of file m_fish.f90.
◆ fish_midgut_shift_items_after_new_ingest()
| elemental subroutine the_fish::fish_midgut_shift_items_after_new_ingest | ( | class(midgut), intent(inout) | this, |
| class(food_item), intent(in), optional | food_item_pass | ||
| ) |
Shift food items in the midgut after a new food item is ingested and therefore food items are shifted in stomach adding the new item.
- Parameters
-
[in] food_item_pass Optional food item that is ingested by the fish (environ::food_item) or any extension class. This food mass is used only to set the reference start mass of the food item.
- The food item is then added to the stack array of the fish stomach. It takes the first place shifting all other food items in sequence.
- The first food item in the stack is then obtained from the input parameter food_item_pass. But note that at this stage, its actual environ::food_item::mass is set to zero, the starting mass food_item_eaten::mass_0 is obtained from the input item's actual mass. The food_item_eaten::time_in_process is set to zero. All this is done via the food_item_eaten::zero() method.
Definition at line 2072 of file m_fish.f90.
◆ appetite_func()
| elemental real(srp) function the_fish::appetite_func | ( | real(srp), intent(in) | mass_relative, |
| real(srp), intent(in) | A, | ||
| real(srp), intent(in) | R | ||
| ) |
Computational backend for the appetite function. Appetite is calculated as.
![]()
where ![]() is the relative filling capacity (range 0 to 1) of the fish stomach,
is the relative filling capacity (range 0 to 1) of the fish stomach, ![]() and
and ![]() are logistic function parameters.
are logistic function parameters.
gnuplot code:
set xrange [0:1] plot 1-1/(1+500000*exp(-20*x))
- Note
- Note that m_ifcmd::save_appetite_function_data() saves the appetite factor data for plotting, testing and analysis.
- Parameters
-
[in] mass_relative The mass of food in the stomach relative to the filling capacity. [in] a Parameters of the logistic function 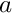 .
. [in] r Parameters of the logistic function 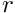 .
.
- Returns
- Returns the appetite factor value that ranges from 0 to 1.
Definition at line 2134 of file m_fish.f90.
◆ fish_appetite_probability_ingestion()
| elemental real(srp) function the_fish::fish_appetite_probability_ingestion | ( | class(fish), intent(in) | this, |
| integer(long), intent(in), optional | time_step | ||
| ) |
Calculate the overall appetite of the fish, which defines the probability to ingest any food item.
- Parameters
-
[in] time_step Time step, optional parameter
Fish appetite at night is near-zero
The overall appetite is a function of individual appetite factors:
- appetite factor for stomach the_fish::stomach::appetite_stomach()
- appetite factor for midgut the_fish::midgut::appetite_midgut()
- appetite factor for energy balance the_fish::fish::appetite_energy()
Version 1 multiplication rule: ![]() probability of a combination of events events.
probability of a combination of events events.
Version 2 maximum/competition rule:
![]()
Definition at line 2150 of file m_fish.f90.
◆ stress_suppress_factor_s()
| pure real(srp) function the_fish::stress_suppress_factor_s | ( | integer, intent(in) | time_step_incr, |
| logical, intent(in), optional | is_force | ||
| ) |
Calculate suppression factor for appetite caused by stress intervention at time time: generic interface is ::stress_factor(). This is the scalar version, vector -based version is stress_suppress_factor_v().
+**
F | ***
| **
+-------*--
^ time_step_incr
A-F - Parameters
-
[in] time_step_incr Time step increment from the moment stress intervention has occurred [in] is_force optional flag to force calculation of the function even if stress is disabled
Definition at line 2213 of file m_fish.f90.
◆ stress_suppress_factor_v()
| pure real(srp) function, dimension(:), allocatable the_fish::stress_suppress_factor_v | ( | integer, dimension(:), intent(in) | time_step_incr, |
| logical, intent(in), optional | is_force | ||
| ) |
Vector-based version of the stress-based suppression function ::stress_factor(). There is a scalar version of the same function stress_suppress_factor_s.
- Parameters
-
[in] time_step_incr Time step increment from the moment stress intervention has occurred [in] is_force optional flag to force calculation of the function even if stress is disabled
Definition at line 2243 of file m_fish.f90.
◆ stress_suppress()
| pure real(srp) function the_fish::stress_suppress | ( | integer(long), intent(in), optional | time_step | ) |
Calculate real stress suppression factor for a specific time step given the configured stress intervention pattern. This is a high-level interface to ::stress_factor().
1** 2**
F | *** | ***
| ** | **
--#-------+-------*----+-----#--*---
^ ^
F=0 +<---># time_over
^
A-F - Parameters
-
[in] time_step Current time step
Definition at line 2285 of file m_fish.f90.
◆ fish_body_mass_current_get()
| elemental real(srp) function the_fish::fish_body_mass_current_get | ( | class(fish_skel), intent(in) | this | ) |
Get the current body mass of the fish. Note that this is the shortcut to obtain the last value from the body mass history array the_fish::fish_skel::history::body_mass_current.
Definition at line 2337 of file m_fish.f90.
◆ fish_appetite_factor_energy_balance()
| elemental real(srp) function the_fish::fish_appetite_factor_energy_balance | ( | class(fish), intent(in) | this | ) |
Calculate appetite factor that depends on the energy balance.
Implementation notes
The energy component of the appetite is defined by the following formula:
![]()
gnuplot> set xrange [-1:1] gnuplot> plot 1 / (1+2.718**(-40*(-x-0.2)) )
Definition at line 2346 of file m_fish.f90.
◆ fish_decide_eat_food_item()
| impure subroutine the_fish::fish_decide_eat_food_item | ( | class(fish), intent(inout) | this, |
| class(food_item), intent(in) | food_item_in, | ||
| logical, intent(out), optional | have_eaten | ||
| ) |
This procedure determines if the fish decides to consume the food item and if yes, ingests it using the ::do_ingest() method. @important This version accepts raw food item. Generic interface linking both raw and pointer versions is ambiguous.
- Note
- This cannot be pure due to random operation
- Parameters
-
[in] food_item_in The food item that is available for decision and consumption. [out] have_eaten Optional indicator showing if the fish have actually eaten the food item (returns TRUE).
Definition at line 2416 of file m_fish.f90.
◆ fish_decide_eat_food_item_ptr()
| impure subroutine the_fish::fish_decide_eat_food_item_ptr | ( | class(fish), intent(inout) | this, |
| class(food_item), intent(in), pointer | food_item_in, | ||
| logical, intent(out), optional | have_eaten | ||
| ) |
This procedure determines if the fish decides to consume the food item and if yes, ingests it using the ::do_ingest() method. @important This version accepts pointer to food item. Generic interface linking both raw and pointer versions is ambiguous.
- Note
- This cannot be pure due to random operation
- Parameters
-
[in] food_item_in The food item that is available for decision and consumption. [out] have_eaten Optional indicator showing if the fish have actually eaten the food item (returns TRUE).
Definition at line 2438 of file m_fish.f90.
◆ fish_activity_baseline_component()
| elemental real(srp) function the_fish::fish_activity_baseline_component | ( | class(fish), intent(in) | this, |
| integer(long), intent(in), optional | time_step | ||
| ) |
Calculate the baseline activity of the fish.
Global baseline locomotor activity pattern (swimming speed) for each time step consists of two components:
- baseline that differs at day ::global_baseline_activity_day and night ::global_baseline_activity_night
- feeding activity increment defined by feeding activity factor ::global_feeding_activity_factor
This is illustrated by the following diagram:
+----+ feeding activity multiplier
| |
+--+ - -+----------------+
| | baseline
-------------------+ +-------------------> activity
-night-------------+-day--------------------+-night-------------> - Parameters
-
[in] time_step Optional time step of the model, default value is commondata::global_time_step
Definition at line 2477 of file m_fish.f90.
◆ fish_activity_appetite_factor()
| elemental real(srp) function the_fish::fish_activity_appetite_factor | ( | class(fish), intent(in) | this | ) |
Calculate activity factor linked to appetite. Appetite factor has a linear relationship with the fish appetite !> ![]() , here
, here ![]() is the maximum value of the appetite activity factor when appetite value is maximum
is the maximum value of the appetite activity factor when appetite value is maximum ![]() . Note that the appetite activity factor works as a multiplier for the baseline actvity the_fish::fish::activity_baseline(), see fish_amr_locomotion_energy_cost().
. Note that the appetite activity factor works as a multiplier for the baseline actvity the_fish::fish::activity_baseline(), see fish_amr_locomotion_energy_cost().
Gnuplot commands for plotting:
set xrange [0:1] set yrange [0:5] plot 0.5*x + 1
Definition at line 2523 of file m_fish.f90.
◆ stomach_adjust()
| elemental real(srp) function the_fish::stomach_adjust | ( | integer, intent(in), optional | target_emptying_time | ) |
Calculate the adjustment factor for the transformation of the stomach transport pattern Time scale commondata::global_transport_pattern_t for a different temperature or fish size.
- Parameters
-
[in] target_emptying_time Optional maximum stomach emptying time for a different target temperature, default value is calculated by stomach_emptying_time().
Definition at line 2534 of file m_fish.f90.
◆ stomach_emptying_time()
| elemental integer function the_fish::stomach_emptying_time | ( | real(srp), intent(in), optional | fish_mass, |
| real(srp), intent(in), optional | temperature | ||
| ) |
Calculate the stomach emptying time for a fish with specific body mass at a specific temperature. Calculation is based on spline interpolation of experimental data.
- Parameters
-
[in] fish_mass Optional fish mass, default value is commondata::global_body_mass [in] temperature Optional temperature, default value is commondata::global_temperature
Definition at line 2563 of file m_fish.f90.
◆ fish_stomach_emptying_time()
| elemental integer function the_fish::fish_stomach_emptying_time | ( | class(stomach), intent(in) | this, |
| real(srp), intent(in), optional | temperature | ||
| ) |
Object-oriented frontend for stomach_emptying_time(): Calculate the stomach emptying time for a fish with specific body mass at a specific temperature. Calculation is based on spline interpolation of experimental data.
- Parameters
-
[in] temperature Optional temperature, if absent use commondata::global_temperature parameter.
Definition at line 2608 of file m_fish.f90.
◆ get_stomach_emptying_matrix_csv()
| impure type(stomach_emptying_pattern) function the_fish::get_stomach_emptying_matrix_csv | ( | character(len=*), intent(in) | csv_file | ) |
Get the stomach emptying time base matrix from a CSV input data file.
File structure:
| 50.00 100.00 300.00 500.00
---------+------------------------------------
5.00 | 24.00 35.00 75.00 100.00
10.00 | 15.00 20.00 50.00 75.00
15.00 | 9.00 15.00 40.00 50.00
20.00 | 5.00 10.00 30.00 35.00
22.00 | 4.00 8.00 29.00 33.00 - Parameters
-
[in] csv_file Input file name
Definition at line 2640 of file m_fish.f90.
Variable Documentation
◆ history_size
| integer, parameter, public the_fish::history_size = 20 |
The size of the history that saves the fish data back in time.
Definition at line 24 of file m_fish.f90.